News
October 26, 2020
Reading between the lines of the human genome
DNA in our cells contains the master copy of instructions for building proteins. When a gene is activated, a part of that DNA sequence is read out and transcribed into a messenger RNA (mRNA) molecule, which can go on to instruct building of protein. But there is also a lot of DNA in the genome that does not code for proteins, so-called non-coding DNA. non-coding DNA includes functional regions, also known as enhancers, which help to turn genes on and off in response to the cell’s protein needs. But we are only just starting to learn about how nuances of non-coding DNA systems can lead to failures in our protein manufacture and even disease.
To map out enhancers in the non-coding genome, scientists have started to use a variety of different techniques, collectively called massively parallel reporter assays, or MPRAs. These methods have the purpose of screening thousands of different DNA sequences in a single experiment to build a picture of non-coding sequences for their enhancer activity. But with many different methods currently being used, it is not clear if these are all giving the same answers.
Writing in Nature Methods an international collaboration of scientists has cross-checked the results from nine different ways of performing MPRAs. They identify some of the more dependable methods and confirm some fundamental assumptions about these tests that had been taken on faith by the scientific community.
Co-first-author of Fumitaka Inoue who worked on the study at the University of California San Francisco and is now an associate professor at Kyoto University says, “The human genome is so extensive that we could never fully characterize all the functional DNA elements with conventional one-by-one reporter assays. MPRA techniques are clearly the solution we need, but now everyone is using different approaches. We cannot take these all results at face value, or compare between different methods, without questioning how the design choices are impacting the outcomes.”
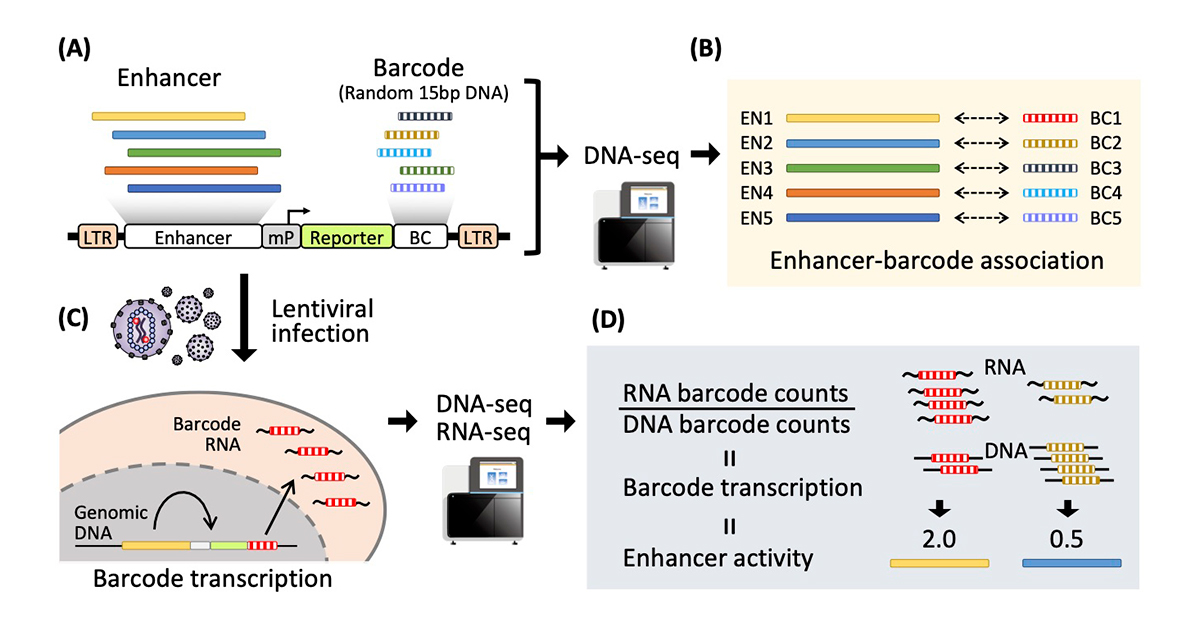
Different recipes for MPRAs blend the same basic parts in slightly different ways. Usually, thousands of enhancer sequences are constructed based on potentially interesting non-coding sequences, which might stimulate a gene expression. Each sequence is attached to a unique DNA sequence, called a barcode.
This DNA construct is delivered into cells, where functional enhancers lead to transcription of the attached barcode into mRNA molecules. By directly measuring the ratio of the DNA and mRNA barcodes from the cells, it is possible to see which enhancers are having the greatest influence on gene expression.
To find the best technique for this experiment the team tested a range of common methods to deliver 2,440 enhancers into liver cells. They looked at different ways of arranging the enhancers and barcodes, and the two common approaches of introducing the sequence into cells, either as a loop of DNA (plasmid) or by infection with a kind of virus (lentivirus). They also duplicated the results of each assay three times to confirm the reliability of each method.
They found that all the assays gave similar readouts but three methods stood out as giving highly reproducible results with a good dynamic range. Among these was the groups own lentivirus-based MPRA method.
Although the results were quite similar between the different methods, they did find some signs that the transcription factors and RNA-binding proteins that get bound by the enhancers influence their activity in different methods. Further testing also clarified the commonly accepted idea that the direction of the enhancer sequence has only a minor influence on activity.
Inoue says, “Our results do suggest a degree of caution in interpreting the results of all MPRAs, as they are all subject to some influence from the assay design. But, at the same time, our work supports the robustness of the MPRA technologies in regard to its accuracy and reproducibility. This knowledge of MPRA design will be important for all researchers working genomics, medical sciences, and evolutionary biology.”
Paper Information
Jason C. Klein, Vikram Agarwal, Fumitaka Inoue, Aidan Keith, Beth Martin, Martin Kircher, Nadav Ahituv and Jay Shendure (2020). A systematic evaluation of the design and context dependencies of massively parallel reporter assays, Nature Methods, DOI: https://doi.org/10.1038/s41592-020-0965-y
Credit: Nano Pico Science
