
Fumitaka Inoue
Co-PI (Bourque-G)
- Position
- Associate Professor
- Research Field
- Genomics, Molecular Developmental Biology
- ORCID
- https://orcid.org/0000-0003-0657-434X
- Personal Website
- https://ashbi.kyoto-u.ac.jp/lab-sites/inoue_lab/
Research Overview
Understanding human regulome that underlies cell differentiation, development and evolution
In our genome, only 1.5% of DNA encodes for proteins and 98% is non-coding. Non-coding DNA includes gene regulatory elements, called “enhancers”. Enhancers respond to inter-/intra-cellular signals or stresses, interact with transcription factors and histone modifications, and regulate gene expression at appropriate time, location and quantity. However, because of the difficulty in identifying and characterizing functional enhancers, regulatory landscape in our genome, or regulome, is still largely unclear.
We have been identifying enhancers that are involved in cell differentiation, development, and evolution using genomic/epigenomic techniques, such as RNA-seq, ChIP-seq, Cut&Tag and ATAC-seq. In addition, we have developed lentivirus-based massively parallel reporter assay (lentiMPRA), a novel technology that enables functional characterization of enhancers in a high-throughput and quantitative manner by using transcribed barcodes (Fig 1). Using these techniques, we have been characterizing thousands of functional enhancers in neural progenitors differentiated from primate pluripotent stem cells. Our laboratory aims to understand human regulome that underlies complex biological phenomena, such as cell differentiation, development and evolution, by applying cutting edge genomic technologies in combination with iPS cell and single-cell technologies.
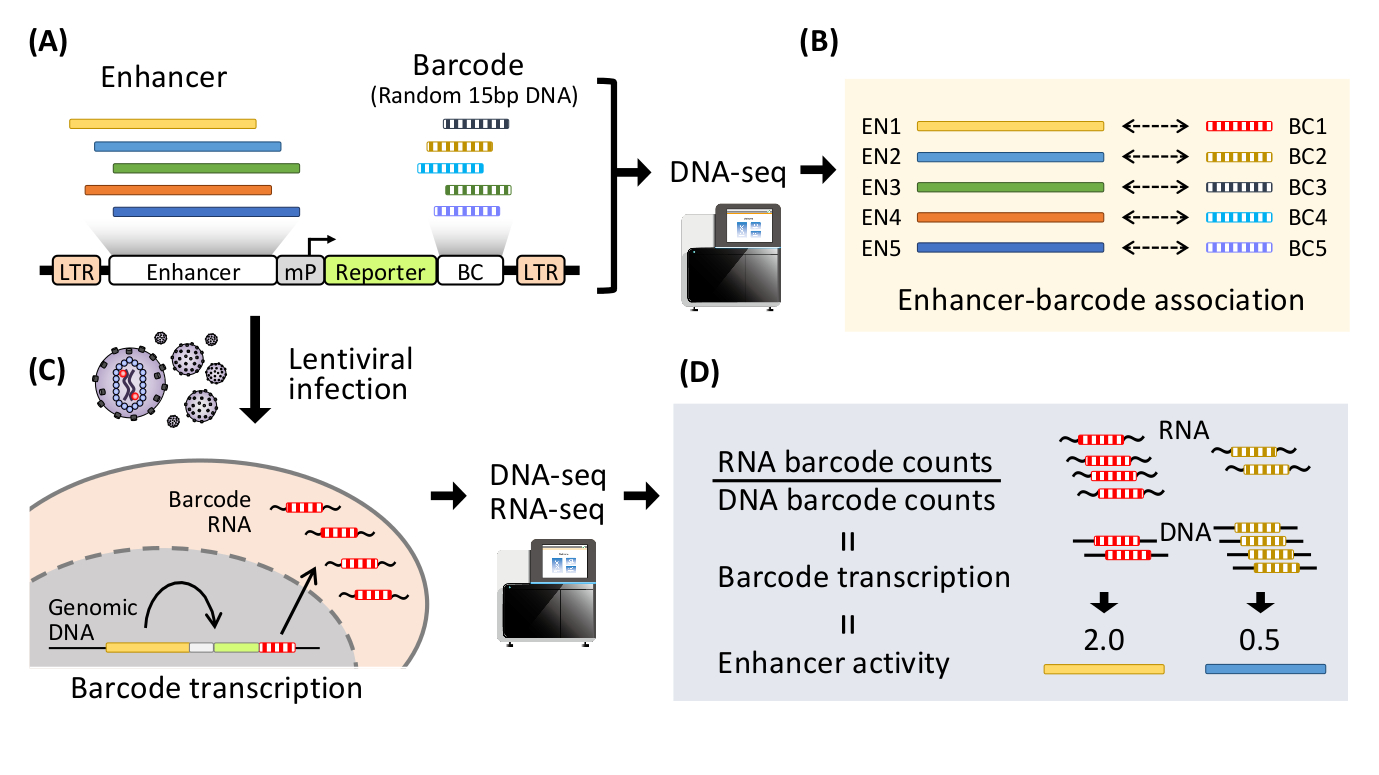
Fig 1: Schematic diagram of lentivirus-based massively parallel reporter assay (lentiMPRA)
(A) Thousands of enhancer candidates are inserted into upstream of a minimal promoter (mP) in the lentivirus vector. Barcodes (BC, random 15bp DNA) are inserted into 3’UTR of a reporter gene to create lentiMPRA library. (B) The lentiMPRA library is sequenced to associate enhancer candidates with barcodes. (C) The lentiMPRA library is infected into the cells of interest via lentivirus. The vector DNA is integrated in the host genome, and barcodes are transcribed under the regulation of the enhancer associated. (D) DNA and RNA from the cells is sequenced to quantify barcode transcription and enhancer activity in a massively parallel manner.
Biography
Fumitaka Inoue obtained his PhD from Saitama University (2008) and undertook postdoctoral training at RIKEN, center for developmental biology (2008-2012). He moved to University of California, San Francisco (2012-2020) as a postdoctoral fellow. He was appointed Associate Professor in 2020 in ASHBi, Kyoto University.
Publications
Kathleen C. Keough, Sean Whalen, Fumitaka Inoue, Pawel F. Przytycki, Tyler Fair, Chengyu Deng, Marilyn Steyer, Hane Ryu, Kerstin Lindblad-Toh, Elinor Karlsson, Zoonomia Consortium, Tomasz Nowakowski, Nadav Ahituv, Alex Pollen, Katherine S. Pollard
Three-dimensional genome rewiring in loci with human accelerated regions
Science 380(6643). (2023)
Sean Whalen*, Fumitaka Inoue*, Hane Ryu*, Tyler Fair, Eirene Markenscoff-Papadimitriou, Kathleen Keough, Martin Kircher, Beth Martin, Beatriz Alvarado, Orry Elor, Dianne Laboy Cintron, Alex Williams, Md. Abul Hassan Samee, Sean Thomas, Robert Krencik, Erik M. Ullian, Arnold Kriegstein, John L. Rubenstein, Jay Shendure, Alex A. Pollen, Nadav Ahituv^, Katherine S. Pollard^
Machine learning dissection of human accelerated regions in primate neurodevelopment
Neuron 111(6), 857-873. (2023)
Anat Kreimer*^, Tal Ashuach*, Fumitaka Inoue*, Alex Khodaverdian, Chengyu Deng, Nir Yosef^, Nadav Ahituv^
Massively parallel reporter perturbation assays uncover temporal regulatory architecture during neural differentiation
Nature communications 13(1), 1504. (2022)
Carly V Weiss*, Lana Harshman*, Fumitaka Inoue, Hunter B Fraser, Dmitri A Petrov^, Nadav Ahituv^, David Gokhman^
The cis-regulatory effects of modern human-specific variants
eLife 10. (2021)
Jason C. Klein*, Vikram Agarwal*, Fumitaka Inoue*, Aidan Keith*, Beth Martin, Martin Kircher, Nadav Ahituv^, Jay Shendure^
A systematic evaluation of the design and context dependencies of massively parallel reporter assays
Nature Methods 17, 1083-1091.(2020)
M. Grace Gordon*, Fumitaka Inoue*^, Beth Martin*, Max Schubach*, Vikram Agarwal, Sean Whalen, Shiyun Feng, Jingjing Zhao, Tal Ashuach, Ryan Ziffra, Anat Kreimer, Ilias Georgakopoulous-Soares, Nir Yosef, Chun Jimmie Ye, Katherine S Pollard, Jay Shendure^, Martin Kircher^, Nadav Ahituv^
lentiMPRA & MPRAflow for high-throughput functional characterization of gene regulatory elements
Nature Protocols 15, 2387-2412. (2020)
Fumitaka Inoue*, Anat Kreimer*, Tal Ashuach, Nadav Ahituv^, Nir Yosef^
Identification and massively parallel characterization of regulatory elements driving neural induction
Cell Stem Cell 25, 713-727. (2019)
Fumitaka Inoue*, Walter Eckalbar*, Yi Wang, Karl K. Murphy, Navneet Matharu, Christian Vaisse^, Nadav Ahituv^
Genomic and epigenomic mapping of leptin-responsive neuronal populations involved in body weight regulation
Nature Metabolism 1, 475-484. (2019)
Fumitaka Inoue*, Martin Kircher*, Beth Martin, Gregory M Cooper, Daniela M Witten, Michael T McManus, Nadav Ahituv^ and Jay Shendure^
A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity
Genome Research 27, 38-52. (2017)
Fumitaka Inoue and Nadav Ahituv
Decoding enhancers using massively parallel reporter assays
Genomics 106, 159-64. (2015)
*co-first authors
^co-corresponding authors
Awards
Best poster award (Runner-up), ENCODE consortium meeting (2018)
Joined
Jul. 1, 2020
