Research & Development
Development of Single-Cell and Single-Molecule Epigenome Analysis Techniques
Gene expression is controlled by various nearby genomic regulatory regions. Variations in the sequences of these regulatory regions are often pivotal factors in human disease. Additionally, genomic sequence differences play a significant role in establishing traits of humans and other organisms during evolution. Therefore, a deep understanding of the function and regulatory mechanisms of genomic regions is crucial in human biology.
The interplay between genomic regulatory regions, including promoter–enhancer interactions, is a hallmark of their functionality. In particular, these interactions between genomic regions should be very dynamic. In response to the dynamics of these interactions, the state of genome expression and the cell state are dynamically and yet robustly controlled. We believe that understanding the relationship between genomic interaction dynamics and gene expression profiles is the key to connecting the nature of genomic DNA with human traits.
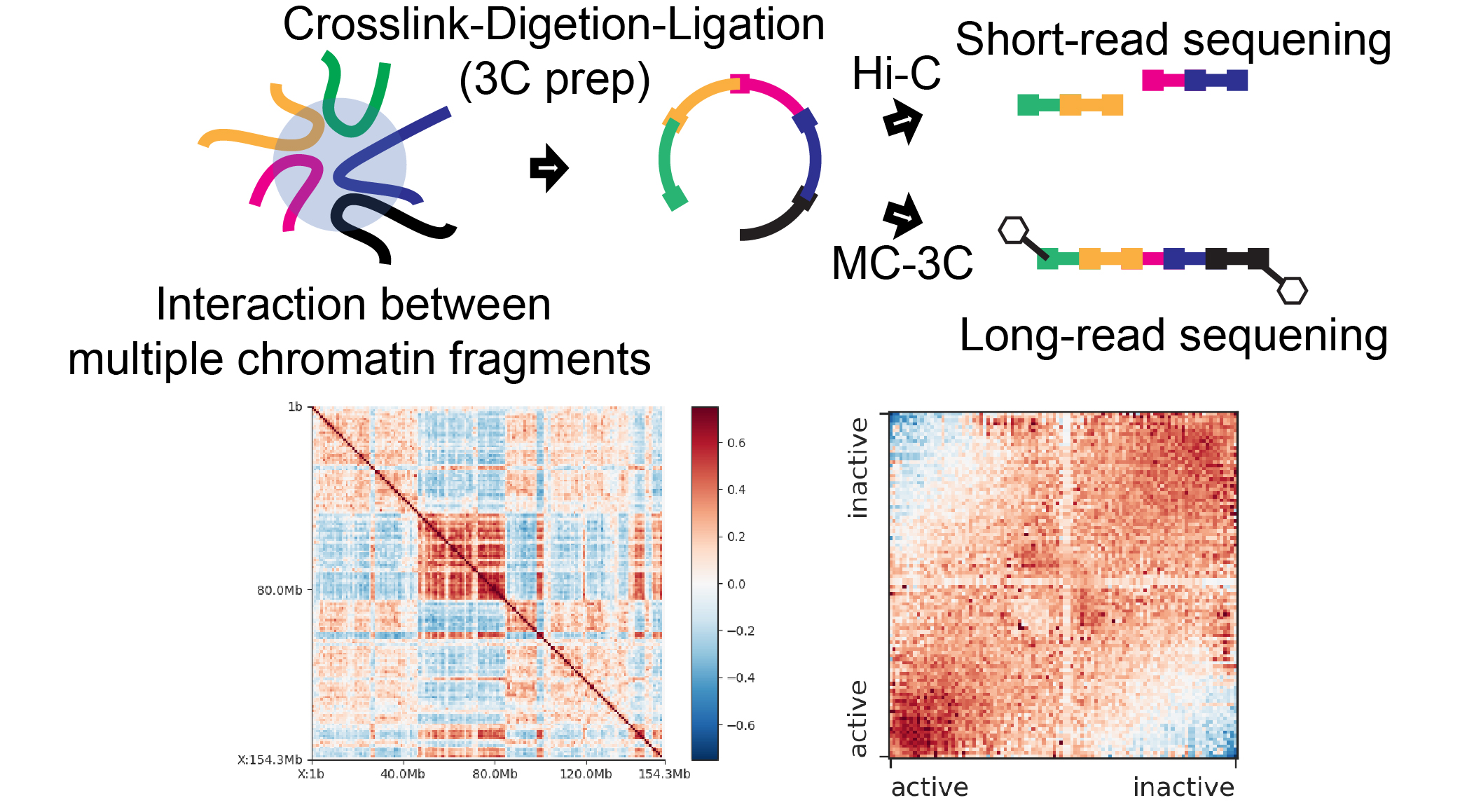
To unravel this, it is essential to analyze the dynamic states of genomic interactions in detail. Although various analytical methods have emerged because of technological innovations, obtaining sufficient information remains a challenge. SignAC focuses on the development and implementation of technologies to reveal genomic interactions at the single-cell and single-molecule levels. Using these technologies, SignAC is also involved in research to analyze how the genome controls the properties of cells and tissues. These studies involve state-of-the-art devices such as next-generation sequencing, long-read sequencers, Fluorescence-Activated Cell Sorting, and automated workstations. We are advancing single-molecule epigenome analysis using long-read sequencers as part of the AMED BINDS project.



Research support
SignAC provides support for equipment use and offers analysis services, along with conducting supportive research using its equipment and expertise.
For example, we have performed DNA methylation analysis using a long-read sequencer, analysis of histone modification patterns using CUT&Tag, and data analysis of single-cell RNA-seq, and the generated data are included in published papers.



(From Yamanaka et al. 2023 Nature)
AMED BINDS
Support for Long-Read Single-Molecule Epigenome Analysis
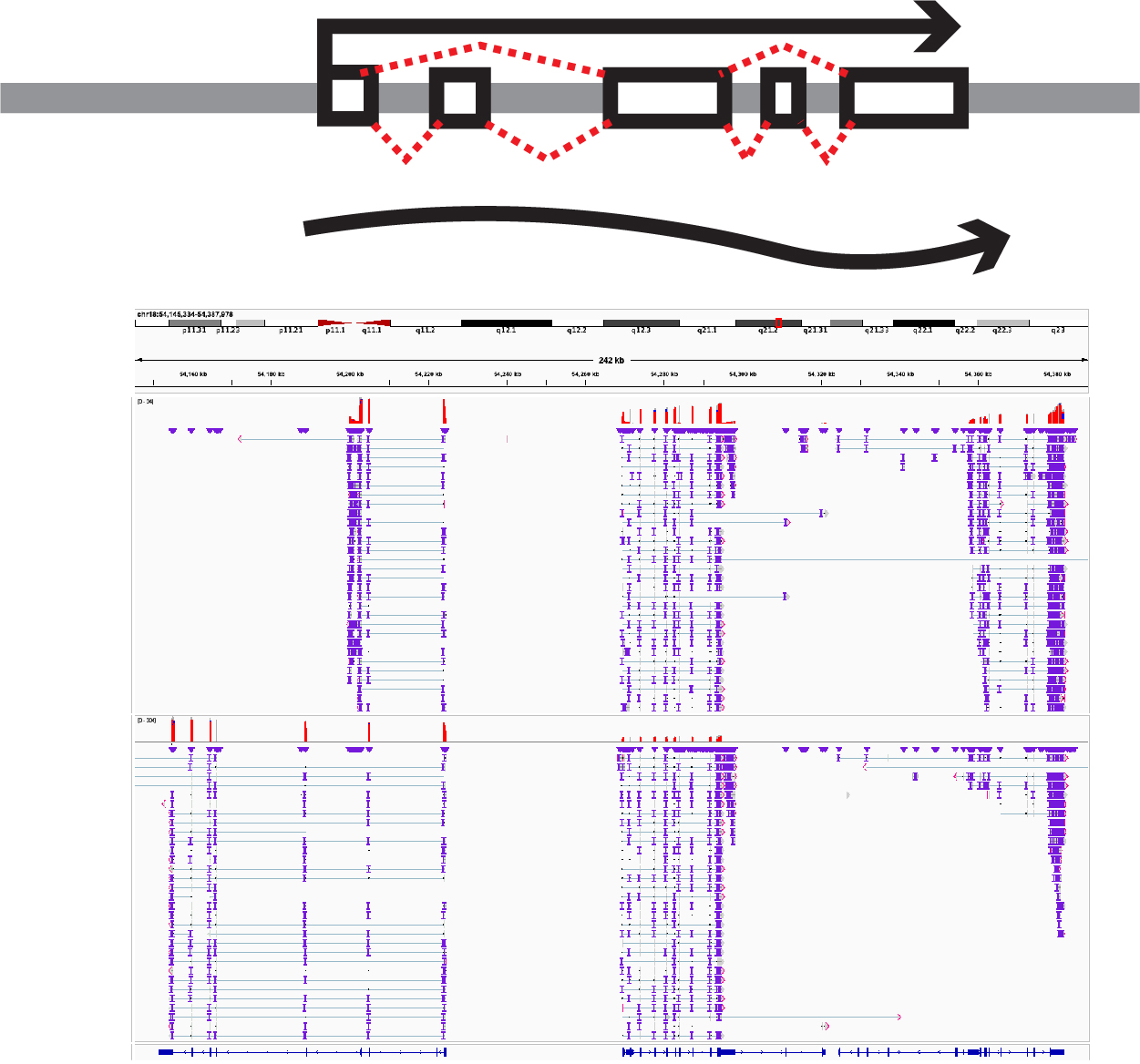
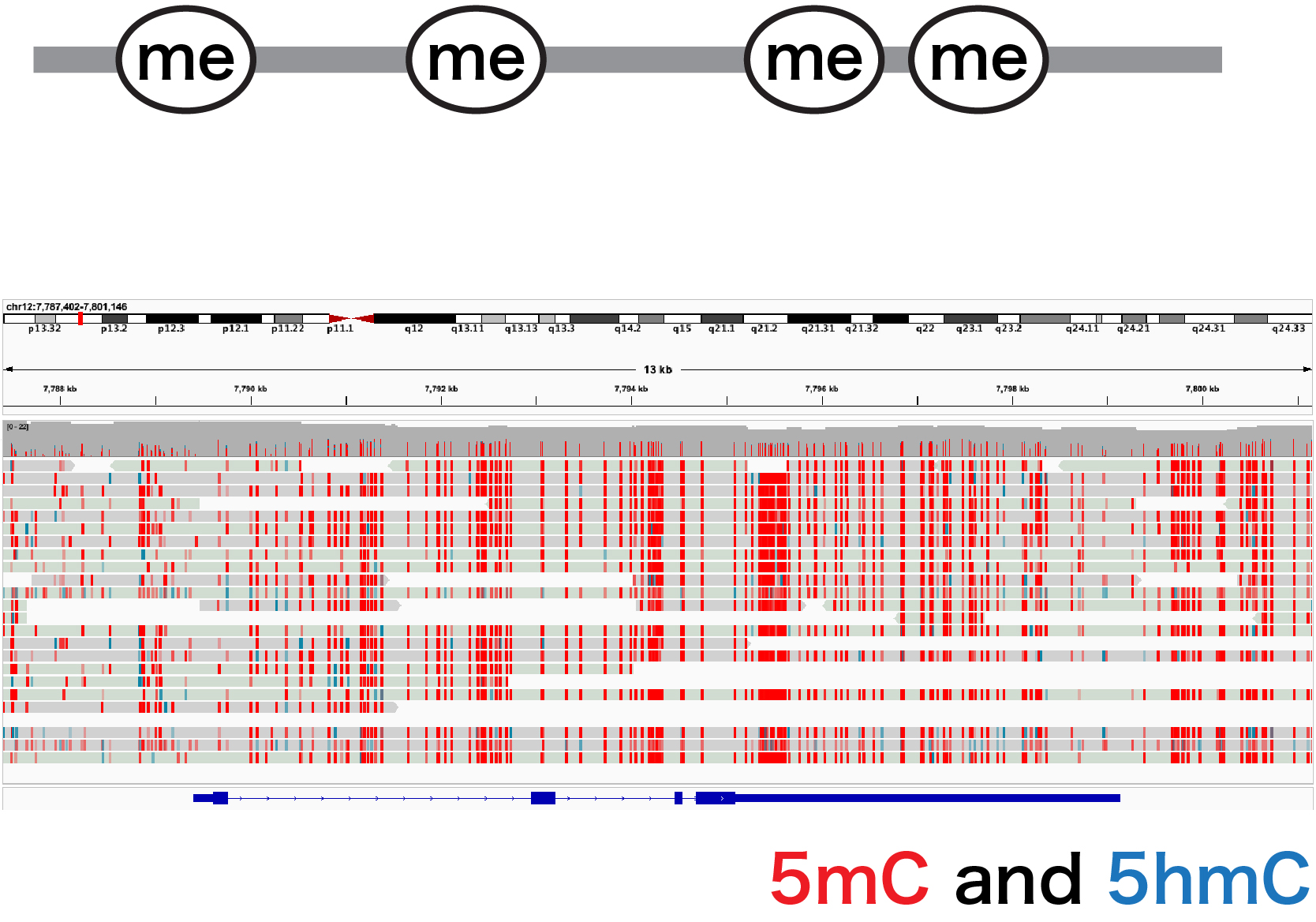
Long-read sequencing is a technology that enables the analysis of DNA sequences and base modification states over tens of thousands of bases single DNA molecules. Through this technology, it is possible to determine the true nature of genomic molecules that function over very long distances, such as through interactions between regulatory regions. SignAC has two large-scale long-read sequencing platforms, ONT’s PromethION24 and PacBio’s Sequel IIe, and conducts support and advanced research for single-molecule epigenome analysis as part of the AMED BINDS project.
Specific support includes full-length transcriptome analysis, long-read methylome analysis, and multicontact chromatin conformation analysis. As a research technological advancement, our primary focus is on the development of these single-cell analysis technologies.