
Kazunori Sunadome
Researcher (T. Yamamoto-G)
- Position
- Assistant Professor
- Research Field
- Cell Biology
Research Overview
Spatial transcriptomics-based analyses of skeletal muscle development and pathologies
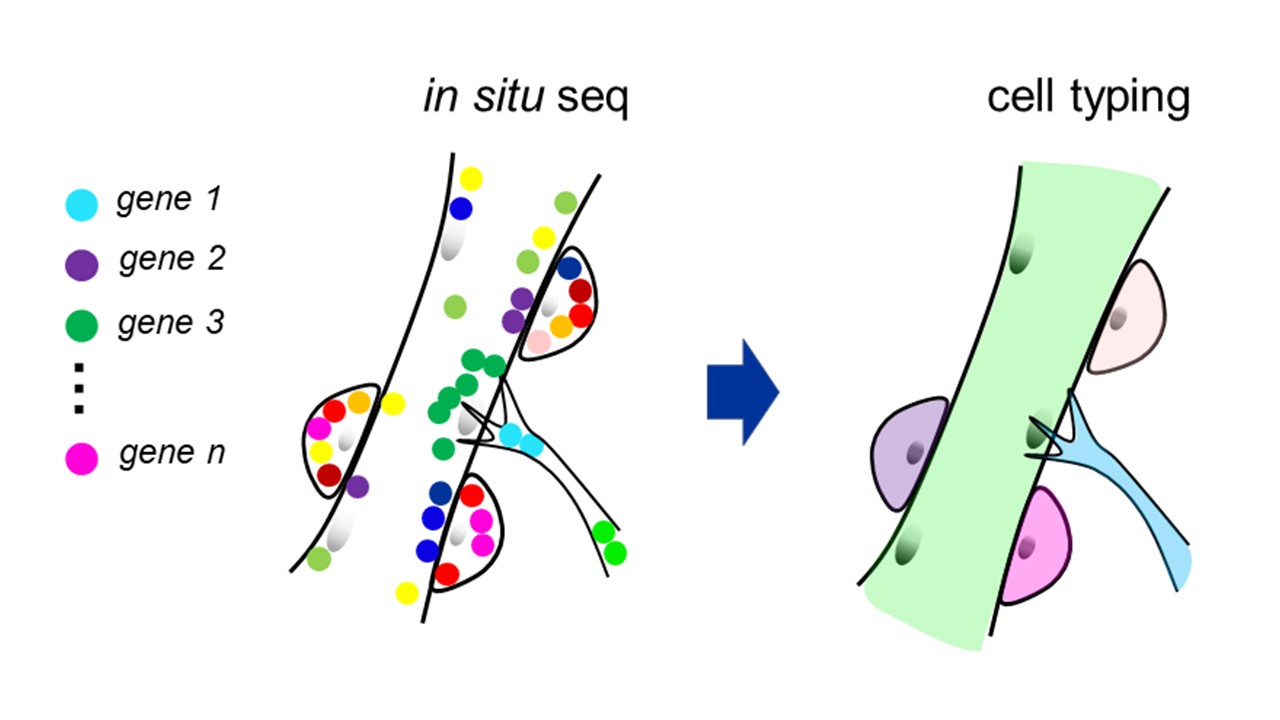
Recent technological advances in in situ imaging of RNAs have allowed us to examine gene expression patterns within individual cells and define cell identity in terms of cell type and state in intact tissues. I am investigating the development and homeostasis of the skeletal muscle through in situ sequencing, which is a spatial transcriptomic method invented by the group of Dr. Mats Nilsson. The skeletal muscle is composed of giant multi-nucleated myofibers surrounded by muscle stem cells (MSCs), which sustain the muscle tissue throughout our lives. I am interested in how myofibers and various cells generate the optimal microenvironment to maintain the regenerative potentials of MSCs and how MSCs are impaired during aging or degenerative diseases. I am also interested in where and how transcripts are localized and translated in lower motor neurons as well as myofibers since neuronal innervation of the muscle is vital for the homeostasis of the tissue. Although humans and rodents presumably share the mechanisms that regulate these processes, human-specific features are also expected considering the characteristic gene regulatory machineries, such as alternative splicing, in human cells. Therefore, I generate artificial tissues from human induced pluripotent stem cells and use them for my analyses. Through these studies, I aim to deepen our understanding of human skeletal muscle physiology and pathologies.
Biography
Kazunori Sunadome obtained his PhD from Kyoto Univeristy (2011), and undertook postdoctoral training in King’s College London (2013-2014) and Karolinska Institute (2015-2020). He joined Dr Takuya Yamamoto’s group from February 2020.
Publications
Zhang Y., Annusver K., Sunadome K., Kameneva P., Edwards S., Lei G., Kasper M., Chagin AS., Adameyko I., Xie M. (2020). Epiphyseal Cartilage Formation Involves Differential Dynamics of Various Cellular Populations During Embryogenesis. Front. Cell Dev. Biol. 8.
Kaucka M., Petersen J., Tesarova M., Szarowska B., Kastriti M.E., Xie M., Kicheva A., Annusver K., Kasper M., Symmons O., Pan L., Spitz F., Kaiser J., Hovorakova M., Zikmund T., Sunadome K., Matise M.P., Wang H., Marklund U., Abdo H., Ernfors P., Maire P., Wurmser M., Chagin A.S., Fried K., Adameyko I. (2018). Signals from the brain and olfactory epithelium control shaping of the mammalian nasal capsule cartilage. Elife 7.
Furlan A., Dyachuk V., Kastriti M.E., Calvo-Enrique L., Abdo H., Hadjab S., Chontorotzea T., Akkuratova N., Usoskin D., Kamenev D., Petersen J., Sunadome K., Memic F., Marklund U., Fried K., Topilko P., Lallemend F., Kharchenko P.V., Ernfors P., Adameyko I. (2017). Multipotent peripheral glial cells generate neuroendocrine cells of the adrenal medulla. Science 357.
Sunadome K., Suzuk, T., Usui M., Ashida Y., and Nishida E. (2014). Antagonism between the master regulators of differentiation ensures the discreteness and robustness of cell fates. Mol. Cell. 54, 526-35.
Sunadome K., Yamamoto T., Ebisuya M., Kondoh K., Sehara-Fujisawa A., and Nishida E. (2011). ERK5 regulates muscle cell fusion through Klf transcription factors. Dev. Cell 20, 192-205
Endo T., Kusakabe M., Sunadome K., Yamamoto T., and Nishida E. (2011). The kinase SGK1 in the endoderm and mesoderm promotes ectodermal survival by down-regulating components of the death-inducing signaling complex. Sci. Signal. 4, ra2.
Kondoh K., Sunadome K., and Nishida E. (2007). Notch signaling suppresses p38 MAPK activity via induction of MKP-1 in myogenesis. J. Biol. Chem. 282, 3058-65.
Research Group
T. Yamamoto GroupJoined
Feb. 1, 2020
