
Bernard de Massy
Research (Saitou-G)
- Position
- Visiting Research Scholar
- Research Field
- Meiosis, Recombination
- ORCID
- https://orcid.org/0000-0002-0950-2758
- Personal Website
- https://www.igh.cnrs.fr/en/research/departments/genome-dynamics/meiose-et-recombinaison
Research Overview
Molecular mechanism and regulation of meiotic recombination
In sexually reproducing species, meiosis allows the formation of haploid gametes from diploid cells. The halving of the DNA content results from a specialized cell cycle, where a single phase of DNA replication is followed by two divisions. The reductional segregation of homologous chromosomes (homologues) at the first meiotic division requires the establishment of connections between homologues. In most species, these connections are established during a long and specialized prophase by reciprocal exchanges between homologues. These exchanges, also called crossing over, result from a highly regulated homologous recombination pathway that drives the recognition and interaction between homologues and the formation of at least one crossing over per homologue pair. Crossovers also generate new allele combinations and thus increase genetic diversity and contribute to genome evolution. The absence of crossover leads to chromosome segregation defects and sterility, and alteration of the meiotic recombination pathway can lead to genome rearrangements and aneuploidy.
Our team is investigating several aspects of the mechanism and regulation of meiotic recombination and its evolutionary implication using the mouse as a model system. Meiotic recombination events are initiated by the formation of DNA double-strand breaks (DSBs, several hundred per nucleus in mice), the repair of which leads to both crossovers and non-crossovers (gene conversion without crossover). The main steps and factors involved in this pathway are evolutionary conserved.
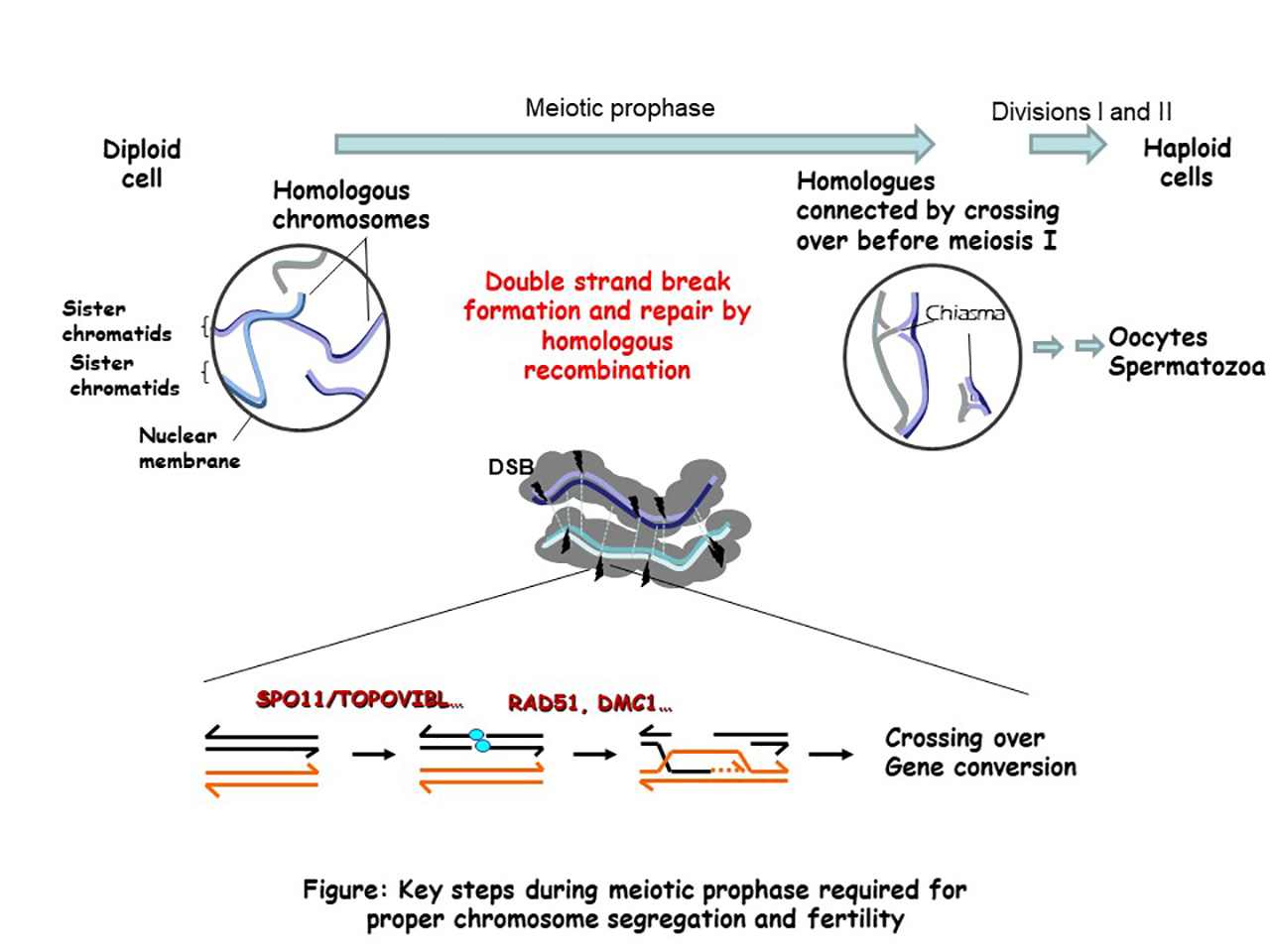
Figure: Key steps during meiotic prophase required for proper chromosome segregation and fertility.
Biography
Bernard de Massy, engineer in agronomy and doctor in microbiology created his own group at the Institute of Human Genetics in 1998 where he developed novel strategies to study in mice the process of homologous recombination during meiosis. His projects have led to major breakthroughs in the field with the identification of several genes essential for this process and specifically for the formation of the programmed DNA double strand breaks that initiate meiotic recombination events.
Publications
Imai*, Y., Biot*, M., Clément, J., Teragaki, M., Urbach, S., Robert, T., Baudat, F., Grey, C., and de Massy, B. (2020) PRDM9 activity depends on HELLS and promotes local 5-hydroxymethylcytosine enrichment. eLife 9. 10.7554/eLife.57117.
Papanikos, F., Clement, J.A.J., Testa, E., Ravindranathan, R., Grey, C., Dereli, I., Bondarieva, A., Valerio-Cabrera, S., Stanzione, M., Schleiffer, A., et al. (2019). Mouse ANKRD31 Regulates Spatiotemporal Patterning of Meiotic Recombination Initiation and Ensures Recombination between X and Y Sex Chromosomes. Mol Cell 74, 1069-1085 e1011. 10.1016/j.molcel.2019.03.022.
Diagouraga, B., Clement, J.A.J., Duret, L., Kadlec, J., de Massy, B., and Baudat, F. (2018). PRDM9 Methyltransferase Activity Is Essential for Meiotic DNA Double-Strand Break Formation at Its Binding Sites. Mol Cell 69, 853-865 e856. 10.1016/j.molcel.2018.01.033. This article has been the subject of a News and Views.
Grey C*, Clement JA*, Buard J, Leblanc B, Gut I, Gut M, Duret L and de Massy B. (2017) In vivo binding of PRDM9 reveals interactions with noncanonical genomic sites. Genome Res. 27, 580-590.
Robert T, Nore A, Brun C, Maffre C, Crimi B, Guichard V, Bourbon HM* and de Massy B. * (2016) The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 351, 943-9. This article has been the subject of a News and Views.
Cole F*, Baudat F*, Grey C, Keeney S*, de Massy B* and Jasin M. * (2014) Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat Genet 46, 1072-80. This article has been the subject of a News and Views.
Wu H, Mathioudakis N, Diagouraga B, Dong A, Dombrovski L, Baudat F, Cusack S, de Massy B* and Kadlec J.* (2013) Molecular Basis for the Regulation of the H3K4 Methyltransferase Activity of PRDM9. Cell Rep 5, 13-20.
Baudat F, Imai Y and de Massy B. (2013) Meiotic recombination in mammals: localization and regulation. Nat Rev Genet 14, 794-806.
Grey C, Barthes P, Chauveau-Le Friec G, Langa F, Baudat F and de Massy B. (2011) Mouse PRDM9 DNA-Binding Specificity Determines Sites of Histone H3 Lysine 4 Trimethylation for Initiation of Meiotic Recombination. PLoS Biol 9, e1001176.
Kumar R, Bourbon HM and de Massy B. (2010) Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev 24, 1266-80. This article has been the subject of a News and Views.
Baudat F*, Buard J*, Grey C*, Fledel-Alon A, Ober C, Przeworski M, Coop G and de Massy B. (2010) PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836-40. This article has been the subject of five News and Views and press conference.
Awards
Price for Discovery by French Academy of Science, 2011
Jules Martin Price by French Academy of Sciences, 2011
EMBO member 2011
Silver medal from CNRS, 2012
ERC Advanced grant in 2013
Price from Fondation Bettencourt-Schueller, Coup d’élan pour la Recherche, 2016
Price Rosen from Fondation pour la Recherche médicale, 2019.
ERC Advanced grant in 2019
