Research


- Functional genomics with single-nucleotide resolution
- Inter- / intra-species comparative functional genomics
Functional genomics with single-nucleotide resolution
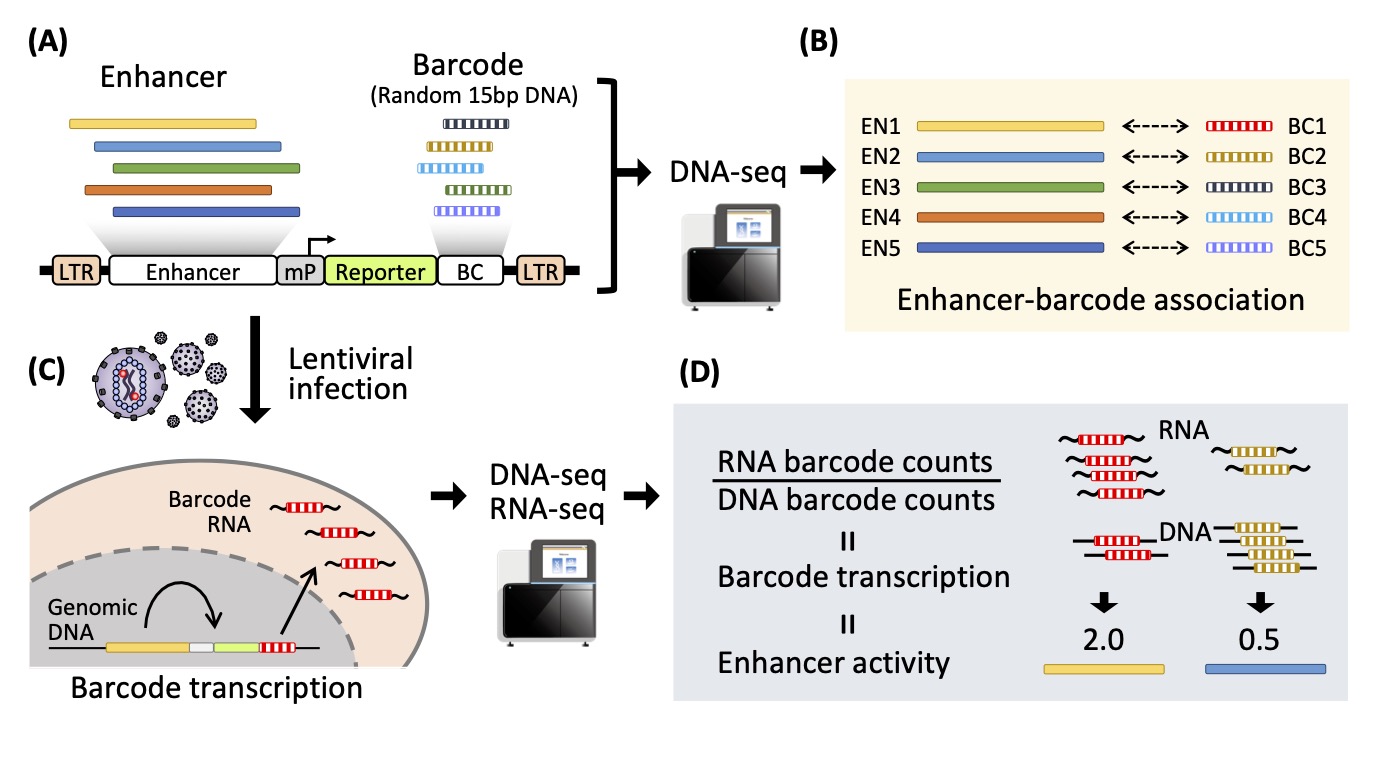
98% of our genome is non-coding, of which function is largely unknown. Recent researches have revealed that the non-coding region includes a variety of “enhancers” that play important role in gene regulation, and their mutations/variations can be a major source of human diseases and evolution. Therefore, deciphering gene regulatory landscape (regulome) is the key to understand human disease and evolution. Inoue has developed lentivirus-based massively parallel reporter assay (lentiMPRA), a novel technology that enables functional characterization of enhancers in a high-throughput and quantitative manner by using transcribed barcodes. We leverage the lentiMPRA and CRISPR activation/interference technologies to dissect enhancer function at single-nucleotide resolution and understand the essence of human regulome.
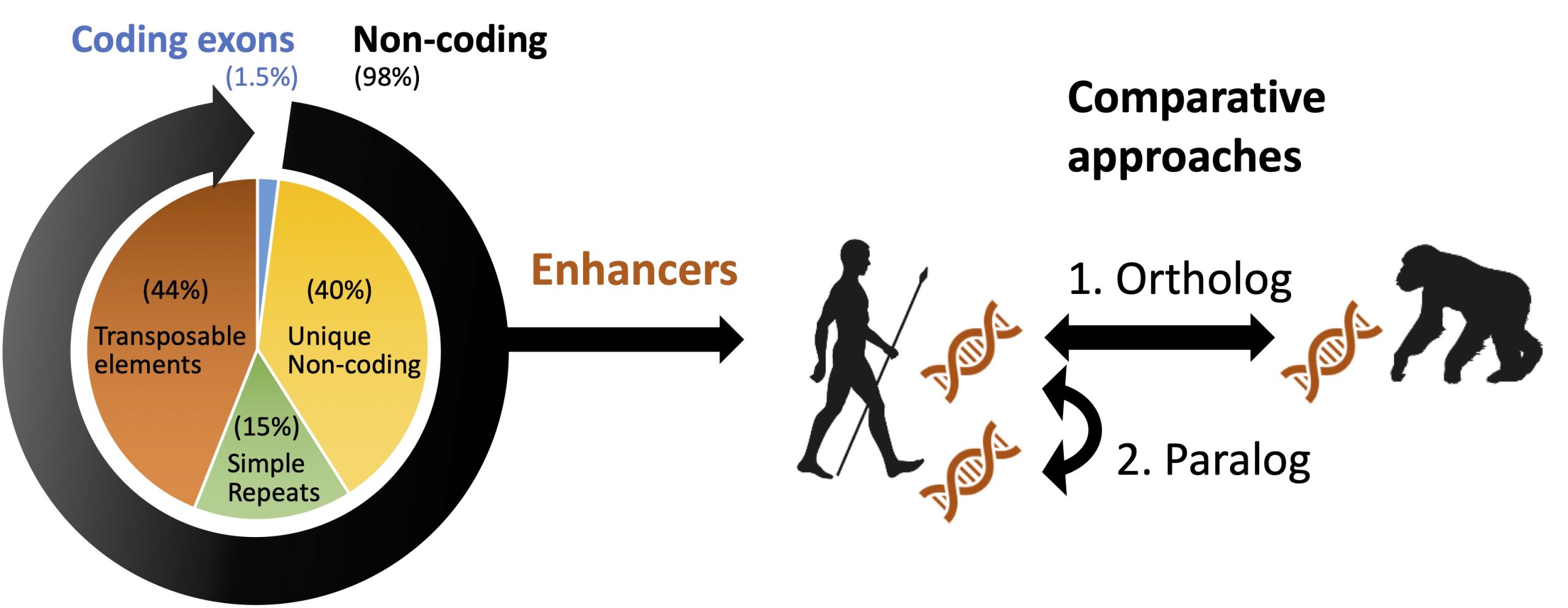
Inter- / intra-species comparative functional genomics
What makes us human? We take following two-directional comparative genomics approaches, aiming to find functional genome regions that evolved in human lineage.
1. Ortholog comparison
By comparing primate genomes/epigenomes (i.e. human, chimpanzee, macaque, etc.), we will identify and analyze enhancer regions that differ among species, and understand what makes us human, and how human genome evolve.
2. Paralog comparison
We especially interested in transposable elements (TEs) in human genome. TEs were integrated and multiplicated in eukaryote genome, and recently elucidated to function as enhancers. Thus, TEs are excellent model for the study of enhancer evolution.
To shape the above approaches, we leverage cutting-edge genomic/epigenomic technologies, such as RNA-seq, ATAC-seq, Cut&Tag and Nanopore long-read DNA-seq.