

Research
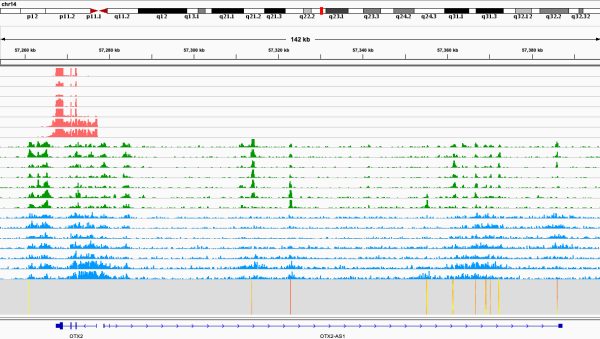
In our genome, only 1.5% of DNA encodes for proteins and 98% is non-coding, of which function is unknown. Inoue laboratory aims to understand human regulome that underlies complex biological phenomena, such as cell differentiation, evolution, and diseases by applying cutting edge genomic/epigenomic technologies.
Pubilications 
- Whalen, S.*, Inoue, F.*, Ryu, H.*, Fair, T., Markenscoff-Papadimitriou, E., Keough, K., Kircher, M., Martin, B., Alvarado, B., Elor, O., Laboy Cintron, D., Williams, A., Hassan Samee, M.A., Thomas, S., Krencik, R., Ullian, E.M., Kriegstein, A., Rubenstein, J.L., Shendure, J., Pollen, A.A., Ahituv, N.,^ Pollard, K.S.^ (2023). Machine learning dissection of human accelerated regions in primate neurodevelopment. Neuron, 111(6), 857–873.DOI
- Kreimer, A.*^, Ashuach, T.*, Inoue, F.*, Khodaverdian, A., Deng, C., Yosef, N.^ & Ahituv, N.^ (2022). Massively parallel reporter perturbation assays uncover temporal regulatory architecture during neural differentiation. Nature Communications, 13(1), 1504. DOI
- Gordon, M. G.*, Inoue, F.*^, Martin, B.*, Schubach, M.*, Agarwal, V., Whalen, S., Feng, S., Zhao, J., Ashuach, T., Ziffra, R., Kreimer, A., Georgakopoulous-Soares, I., Yosef, N., Ye, C. J., Pollard, K. S., Shendure, J.^, Kircher, M.^ & Ahituv, N.^ (2020). lentiMPRA and MPRAflow for high-throughput functional characterization of gene regulatory elements. Nature Protocols, 15(8), 2387–2412. DOI
News
 Recruit
Recruit
We are looking for motivated grad students and postdocs!
Please feel free to contact us.
(inoue.fumitaka.7a☆kyoto-u.ac.jp)replace ☆ to @ and send




