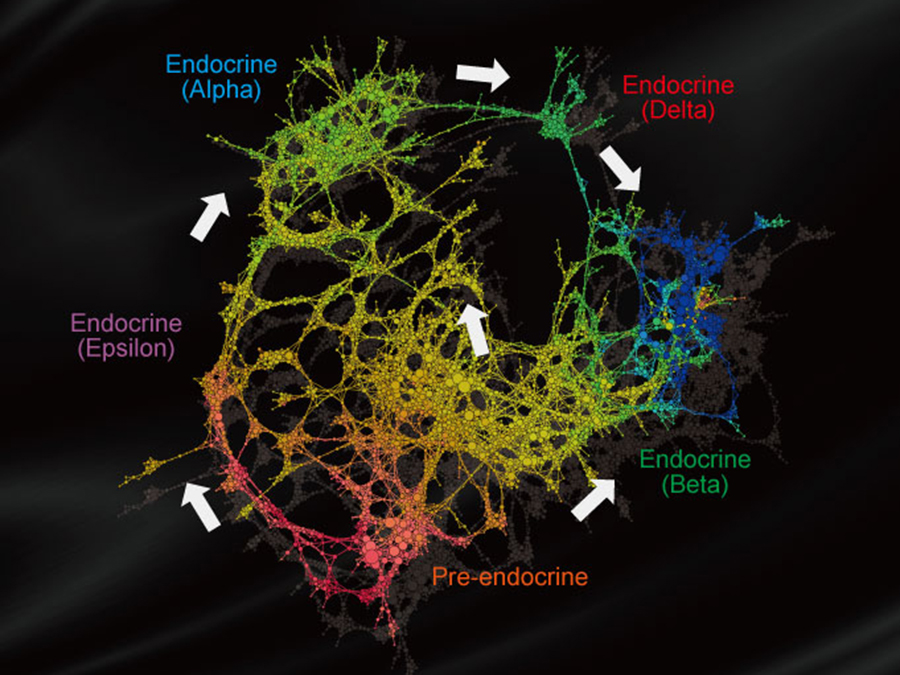
Detecting topology and flow of cell differentiation
Kyoto scientists develop a new trajectory inference method for single-cell RNA-seq data.
Big data has recently become far more available for collection, quickly overwhelming traditional data analysis methods, and the case of single-cell RNA-sequencing (scRNA-seq) is no exception. It is far from obvious how to obtain useful information from data with such a sudden increase in complexity. To this end, rather than blindly attempting to apply existing methods to this new data, we should first describe what structures users want and then design a data analysis method that reflects those structures. Within this century, the field of topological data analysis (TDA), which can detect the “shape” (topology) of data, has been developed and used in a wide range of fields. In the life sciences, Mapper, one of the representative methods in TDA, has been applied to scRNA-seq data to infer a cell differentiation trajectory. Although Mapper can detect the “shape” of cell differentiations, there still remains a problem in that it cannot characterize the “flow” that reflects the time evolution of gene expressions.
Now, the Hiraoka group from the Kyoto University Institute for Advanced Study of Human Biology (WPI-ASHBi) has developed a new mathematical method, V-Mapper (velocity Mapper), that can detect the both “shape” and “flow” of cell differentiation, enabling us to visualize complex structures of cell differentiation from scRNA-seq data.
The lead author Yusuke Imoto said that “V-Mapper can clarify complex cell differentiation mechanisms including topological structures such as the cyclicity and plasticity.”
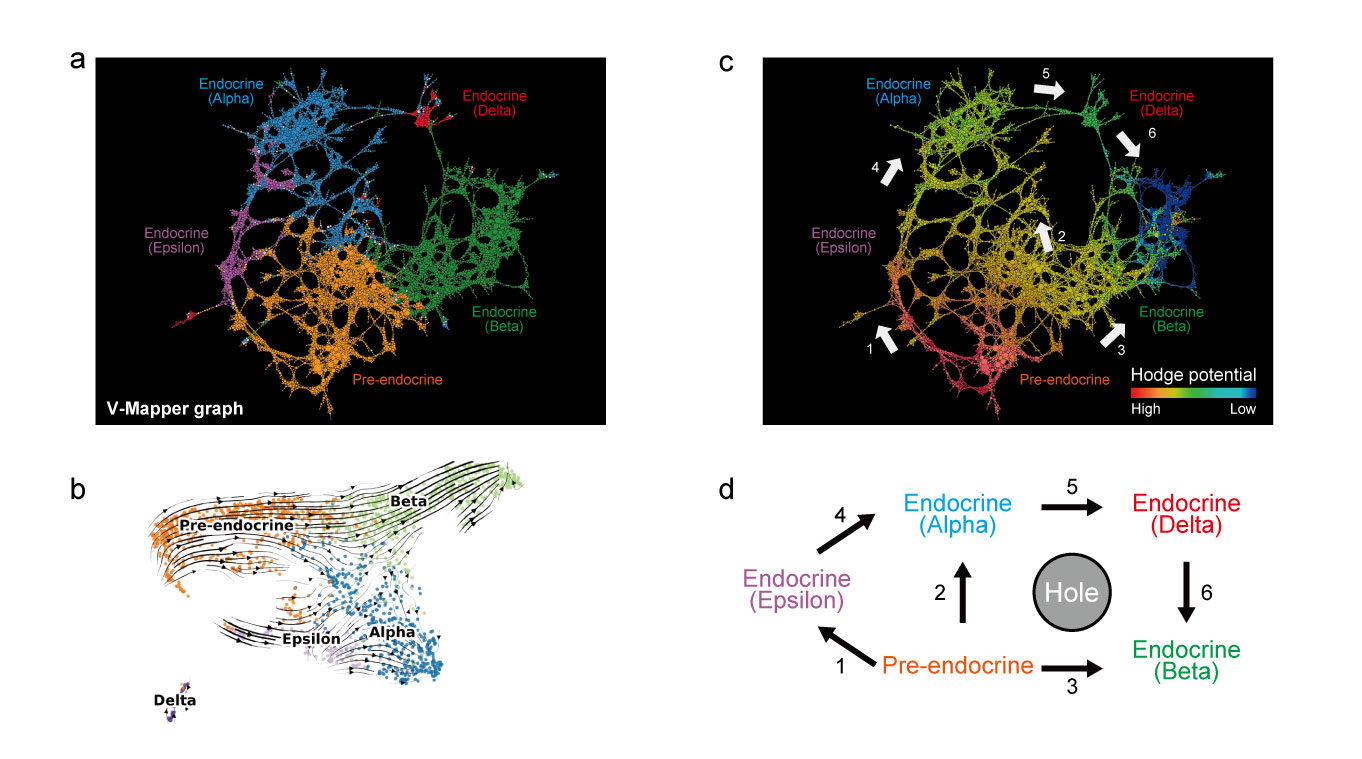
To realize simultaneous visualization of shape and flow, the authors focused on Mapper and RNA velocity, a velocity inference method of gene expression for scRNA-seq data. V-Mapper represents the “shape” and “flow” as a directed graph by embedding a velocity field on the output graph of Mapper. Furthermore, they showed that the graph Hodge decomposition could extract a gradient flow on the V-Mapper graph which reflects a global cell differentiation trajectory from a complex flow structure.
As a biological application, this group applied V-Mapper to scRNA-seq data of mouse pancreatic endocrine cells and successfully detected a complex cell differentiation structure of endocrine cells, which appears in the later stage.
ASHBi vice director Yasuaki Hiraoka comments, “we believe that the scRNA-seq data analysis with V-Mapper will contribute to clinical research such as increasing the efficiency of differentiation induction in regenerative medicine.”
Paper Information
Yusuke Imoto and Yasuaki Hiraoka, (2023). V-Mapper: topological data analysis for high-dimensional data with velocity. Nonlinear Theory and Its Applications (NOLTA), DOI https://doi.org/10.1587/nolta.14.92
Reference
V-Mapper’s Python code is available on GitHub ( https://github.com/yusuke-imoto-lab/V-Mapper )