
Takuya Yamamoto
| Position | Professor |
|---|---|
| Group name | T. Yamamoto Group |
| Research Field | Molecular Biology, Bioinformatics |
Research Overview
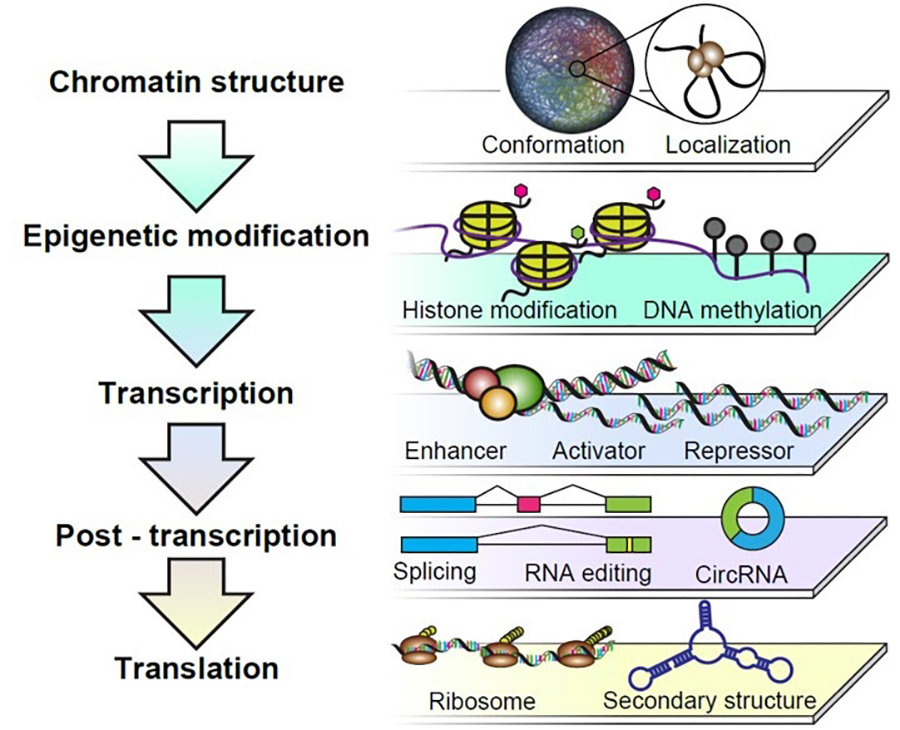
In order to achieve a comprehensive understanding of the regulatory mechanisms underlying gene expressions, it is necessary to investigate multiple regulatory hierarchies including higher-order genomic structures, epigenetic regulation, transcriptional regulation, post-transcriptional regulation and translational regulation (Figure 1). Comprehensive understanding further involves elucidating the intracellular DNA-RNA networks in gene expressions by unveiling relationships between the regulatory hierarchies through the integrative analysis of multiple omics data sets. For this purpose, by taking advantage of molecular biology, biochemistry, cell biology, genetics, genome-wide analyses and bioinformatics techniques, this research group aims to develop an advanced system to investigate a series of regulatory mechanisms that process genomic information into protein synthesis. Ultimately, this system will be used to understand how different phenotypes emerge among different species despite similarities in DNA, RNA, proteins and other cellular infrastructure. Especially, by focusing on the processes responsible for somatic cell reprogramming, cellular differentiation, and organism aging, we have studied the molecular regulatory mechanisms underlying cell fate conversion processes.
This is also responsible for the single-cell genome analysis core, which supports all research at this institute, by providing and developing various genome-wide analytical approaches at the single-cell level.
Publications
Shibata, H., Komura, S., Yamada, Y., Sankoda, N., Tanaka, A., Ukai, T., Kabata, M., Sakurai, S., Kuze, B., Woltjen, K., Haga, H., Ito, Y., Kawaguchi, Y., Yamamoto, T. and Yamada, Y. In vivo reprogramming drives Kras-induced cancer development. Nature Communications 9, 2081 (2018)
Ikeda, H., Sone, M., Yamanaka, S. and Yamamoto T. Structural and spatial chromatin features at developmental gene loci in human pluripotent stem cells. Nature Communications 8, 1616(2017)
Yagi, M., Kishigami, S., Tanaka, A., Semi, K., Mizutani, E., Wakayama, S., Wakayama, T., *Yamamoto, T. and *Yamada, Y. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature 548, 224-227 (2017) *Co-correspondence
Sone, M., Morone, N., Nakamura, T., Tanaka, A., Okita, K., Woltjen, K., Nakagawa, M., Heuser, JE., Yamada, Y., Yamanaka, S. and Yamamoto, T. Hybrid Cellular Metabolism Coordinated by Zic3 and Esrrb Synergistically Enhances Induction of Naive Pluripotency. Cell Metabolism 25, 1103-1117 (2017)
Nakamura, T., Okamoto, I., Sasaki, K., Yabuta, Y., Iwatani, C., Tsuchiya, H., Seita, Y., Nakamura, S., Yamamoto, T. and Saitou, M. A developmental coordinate of pluripotency among mice, monkeys, and humans. Nature 537, 57-62 (2016)


