
Fumitaka Inoue
| Position | Associate Professor |
|---|---|
| Group name | Bourque Group |
| Research Field | Genomics, Molecular Biology |
| Awards | Best poster award (Runner-up), ENCODE consortium meeting (2018) |
| ORCID | https://orcid.org/0000-0003-0657-434X |
| Researchmap | https://researchmap.jp/fumitakainoue?lang=en |
| Personal Website | https://sites.google.com/view/inouelabwebsite/home |
| Joined | Jul. 1, 2020 |
Research Overview
Understanding human regulome that underlies cell differentiation, development and evolution
In our genome, only 1.5% of DNA encodes for proteins and 98% is non-coding. Non-coding DNA includes gene regulatory elements, called “enhancers”. Enhancers respond to inter-/intra-cellular signals or stresses, interact with transcription factors and histone modifications, and regulate gene expression at appropriate time, location and quantity. However, because of the difficulty in identifying and characterizing functional enhancers, regulatory landscape in our genome, or regulome, is still largely unclear.
We have been identifying enhancers that are involved in cell differentiation, development, and evolution using genomic/epigenomic techniques, such as RNA-seq, ChIP-seq, Cut&Tag and ATAC-seq. In addition, we have developed lentivirus-based massively parallel reporter assay (lentiMPRA), a novel technology that enables functional characterization of enhancers in a high-throughput and quantitative manner by using transcribed barcodes (Fig 1). Using these techniques, we have been characterizing thousands of functional enhancers in neural progenitors differentiated from primate pluripotent stem cells. Our laboratory aims to understand human regulome that underlies complex biological phenomena, such as cell differentiation, development and evolution, by applying cutting edge genomic technologies in combination with iPS cell and single-cell technologies.
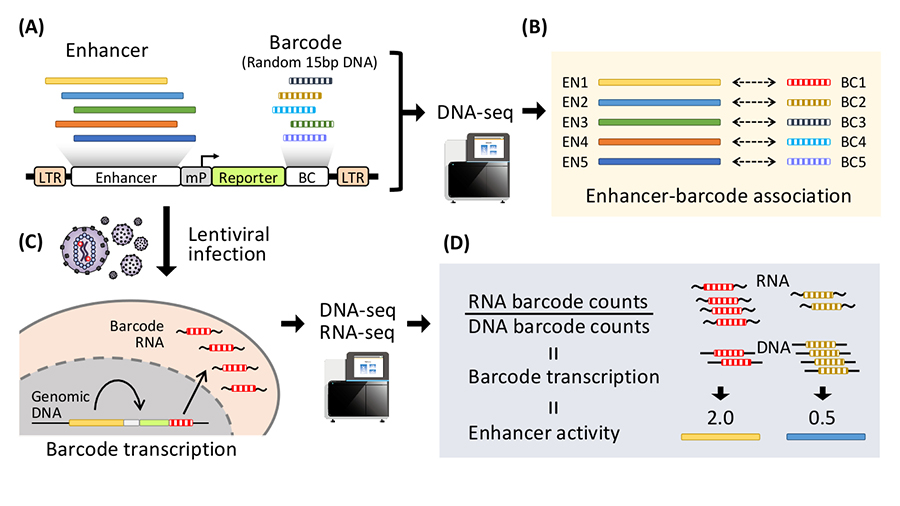
(A) Thousands of enhancer candidates are inserted into upstream of a minimal promoter (mP) in the lentivirus vector. Barcodes (BC, random 15bp DNA) are inserted into 3’UTR of a reporter gene to create lentiMPRA library. (B) The lentiMPRA library is sequenced to associate enhancer candidates with barcodes. (C) The lentiMPRA library is infected into the cells of interest via lentivirus. The vector DNA is integrated in the host genome, and barcodes are transcribed under the regulation of the enhancer associated. (D) DNA and RNA from the cells is sequenced to quantify barcode transcription and enhancer activity in a massively parallel manner.
Biography
Fumitaka Inoue obtained his PhD from Saitama University (2008) and undertook postdoctoral training at RIKEN, center for developmental biology (2008-2012). He moved to University of California, San Francisco (2012-2020) as a postdoctoral fellow. He was appointed Associate Professor in 2020 in ASHBi, Kyoto University.
Publications
Zhang, Z., Georgakopoulos-Soares, I., Bourque, G., Ahituv, N.^, Inoue, F.^ (2026). Simultaneous epigenomic profiling and regulatory activity measurement using e2MPRA. Nature Communications (in press). https://doi.org/10.1038/s41467-026-68422-3
Agarwal, V.*, Inoue, F.*, Schubach, M., Penzar, D., Martin, B.K., Dash, P.M., Keukeleire, P., Zhang, Z., Sohota, A., Zhao, J., Georgakopoulos-Soares, I., Noble, W.S., Yardımcı, G.G., Kulakovskiy, I. V., Kircher, M., Shendure, J., & Ahituv, N. (2025). Massively parallel characterization of transcriptional regulatory elements. Nature, 639, 411–420. https://doi.org/10.1038/s41586-024-08430-9
Chen, X.^, Zhang, Z., Yan, Y., Goubert, C., Bourque, G.^, & Inoue, F.^ (2025). A phylogenetic approach uncovers cryptic endogenous retrovirus subfamilies in the primate lineage. Science Advances, 11. https://doi.org/10.1126/sciadv.ads9164
Bonev, B., Gonçalo, C.-B., Chen, F., Codeluppi, S., Corces, M.R., Fan, J., Heiman, M., Harris, K., Inoue, F., Kellis, M., Levine, A., Lotfollahi, M., Luo, C., Maynard, K.R., Nitzan, M., Ramani, V., Satija, R., Schirmer, L., Shen, Y., Sun, N., Green, G.S., Theis, F., Wang, X., Welch, J.D., Gokce, O., Konopka, G., Liddelow, S., Macosko, E., Bayraktar, O., Habib, N., & Nowakowski, T.J. (2024). Opportunities and challenges of single-cell and spatially resolved genomics methods for neuroscience discovery. Nature Neuroscience, 27, 2292–2309. https://doi.org/10.1038/s41593-024-01806-0
Wang, Q.S., Hasegawa, T., Namkoong, H., Saiki, R., Edahiro, R., Sonehara, K., Tanaka, H., Azekawa, S., Chubachi, S., Takahashi, Y., Sakaue, S., Namba, S., Yamamoto, K., Shiraishi, Y., Chiba, K., Tanaka, H., Makishima, H., Nannya, Y., Zhang, Z., Tsujikawa, R., Koike, R., Takano, T., Ishii, M., Kimura, A., Inoue, F., Kanai, T., Fukunaga, K., Ogawa, S., Imoto, S., Miyano, S., & Okada, Y.; Japan COVID-19 Task Force. (2024). Statistically and functionally fine-mapped blood eQTLs and pQTLs from 1,405 humans reveal distinct regulation patterns and disease relevance. Nature Genetics, 56, 2054–2067. https://doi.org/10.1038/s41588-024-01896-3
Deng, C., Whalen, S., Steyert, M., Ziffra, R., Przytycki, P.F., Inoue, F., Pereira, D.A., Capauto, D., Norton, S., Vaccarino, F.M., Pollen, A.A., Nowakowski, T.J., Ahituv, N., & Pollard, K.S. (2024). Massively parallel characterization of regulatory elements in the developing human cortex. Science, 384. https://doi.org/10.1126/science.adh0559
Feng, Y., Xie, N., Inoue, F., Fan, S., Saskin, J., Zhang, C., Zhang, F., Hansen, M.E.B., Nyambo, T., Mpoloka, S.W., Mokone, G.G., Fokunang, C., Belay, G., Njamnshi, A.K., Marks, M.S., Oancea, E., Ahituv, N., & Tishkoff, S.A. (2024). Integrative functional genomic analyses identify genetic variants influencing skin pigmentation in Africans. Nature Genetics, 56, 258–272. https://doi.org/10.1038/s41588-023-01626-1
Whalen, S.*, Inoue, F.*, Ryu, H., Fair, T., Markenscoff-Papadimitriou, E., Keough, K., Kircher, M., Martin, B., Alvarado, B., Elor, O., Laboy Cintron, D., Williams, A., Hassan Samee, Md.A., Thomas, S., Krencik, R., Ullian, E.M., Kriegstein, A., Rubenstein, J.L., Shendure, J., Pollen, A.A., Ahituv, N., & Pollard, K.S. (2023). Machine learning dissection of human accelerated regions in primate neurodevelopment. Neuron, 111, 857–873.e8. https://doi.org/10.1016/j.neuron.2022.12.026
Keough, K.C., Whalen, S., Inoue, F., Przytycki, P.F., Fair, T., Deng, C., Steyert, M., Ryu, H., Lindblad-Toh, K., Karlsson, E., Nowakowski, T., Ahituv, N., Pollen, A., & Pollard, K.S. (2023). Three-dimensional genome rewiring in loci with human accelerated regions. Science, 380. https://doi.org/10.1126/science.abm1696
Kreimer, A.*, Ashuach, T.*, Inoue, F.*, Khodaverdian, A., Deng, C., Yosef, N., & Ahituv, N. (2022). Massively parallel reporter perturbation assays uncover temporal regulatory architecture during neural differentiation. Nature Communications, 13, 1504. https://doi.org/10.1038/s41467-022-28659-0
*co-first authors
^co-corresponding authors


