
Cantas Alev
| Position | Professor |
|---|---|
| Group name | Alev Group |
| Research Field | Developmental Biology |
| ORCID | https://orcid.org/0000-0002-4879-8782 |
| Researchmap | https://researchmap.jp/alev.cantas?lang=en |
| Joined | Jul. 1, 2019 |
Research Overview
Research in the Alev Lab is driven by the intrinsic curiosity about the very beginnings of human development: how does a single cell give rise to the complexity and wonder of human life?
To understand how we, as human beings, came to be, we are reconstructing embryonic development in the lab, recapitulating key aspects of early development. Through a multidisciplinary approach that brings together synthetic biology, developmental engineering, and omics-based analysis, we aim to create robust, self-organizing, and precisely controllable human embryo models as well as complex tissue models across multiple organ systems. These models will advance our fundamental understanding of human and non-human primate embryogenesis and evolution, while also providing powerful tools for drug discovery, toxicity testing, and the development of new human disease models.
Understanding Early Embryonic Development in Primates
The earliest stages of human development remain some of the most mysterious. While IVF technologies have revealed much about pre-implantation embryos, what happens after implantation is still largely hidden from view. This is the phase when the embryo undergoes dramatic changes: establishing body axes, forming germ layers, and initiating tissue and organ development. But in humans and non-human primates, these stages are exceptionally difficult to study due to ethical and technical constraints, leaving major gaps in our understanding of early development and disease.
At the Alev Lab, we are working to bridge those gaps. Using pluripotent stem cells (PSC)-based model systems, we reconstitute and analyze key developmental events in vitro. This strategy has already yielded important advances in our lab, including the successful recapitulation of the human segmentation clock (Matsuda, Yamanaka et al., Nature, 2020; Matsuda et al., Science, 2020) and the in vitro reconstruction of human somitogenesis (Yamanaka, Hamidi et al., Nature, 2023). We are now working to improve these models, aiming to construct truly robust, self-organizing human embryo models, where spatial and temporal patterning can be precisely controlled, developmental outcomes are reproducible, and large-scale generation is feasible. These advanced models will allow us to unravel the molecular mechanisms that govern species-specific spatial and temporal regulation of complex, multi-germ layer morphogenesis, ultimately illuminating one of life’s most extraordinary transformations.
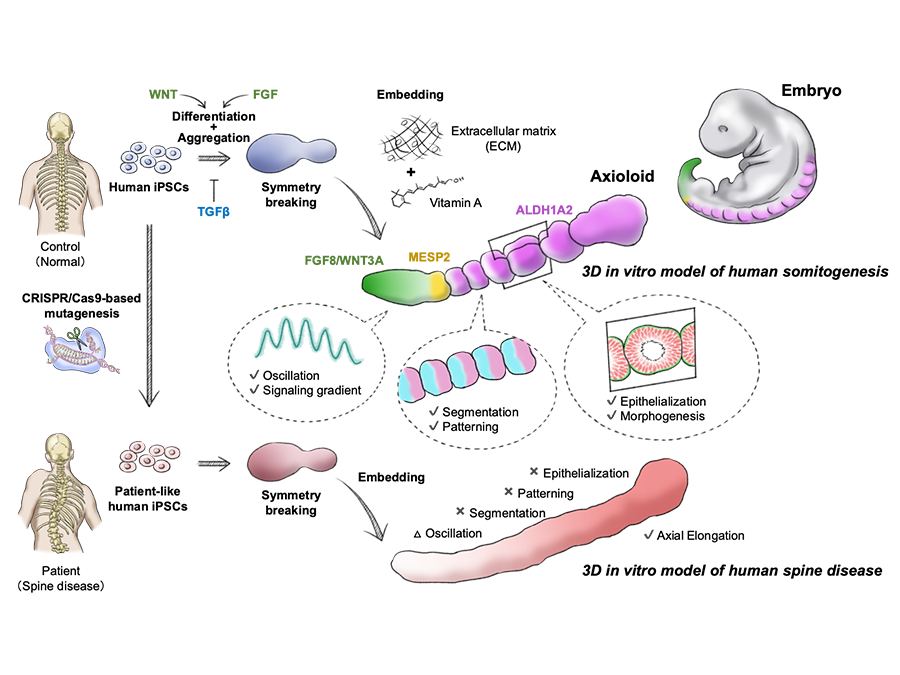
Decoding Axial Skeleton Development and Disease
Many serious conditions affecting the spine and body axis, such as congenital scoliosis and vertebral segmentation defects, can be traced back to the earliest stages of embryogenesis. Our lab is particularly interested in uncovering how these diseases arise and progress during the formation of the axial skeleton. We use PSC-based disease models that mirror early human development under both normal and disease conditions, and perform comprehensive molecular and functional analyses at multiple levels. With this approach, we aim to untangle the complex interplay of genetic, environmental, and epigenetic factors that shape early development and ultimately drive disease.
A key objective of our research is to identify and validate potential development- and disease-associated coding and non-coding genomic regions, as well as epigenetic signatures unique to the human genome. By tracing the roots of disease to the earliest moments of life, our work aims to reveal not only how things go wrong, but also why. Ultimately, we hope these insights will guide the development of new strategies in basic research, disease prevention, and potential therapeutic interventions.
Deciphering Primate Development and Evolution
We are also interested in understanding what makes humans unique. While model organisms such as chick, mouse, and zebrafish have provided valuable insights, species-specific differences in developmental timing, organ size, and tissue complexity make it difficult to fully translate these findings to human biology. Yet, ethical and legal restrictions surrounding human embryo research, as well as significant anatomical and technical challenges, have made it extremely difficult to study early embryonic development directly in humans or other primates.
At the Alev Lab, we tackle this by establishing human and non-human primate PSC models. Combined with multi-omics profiling, we compare human and non-human primate development to identify unique features of human biology. These platforms not only reveal evolutionary differences but can also be adapted to study other developmental processes and organ systems. In addition to human and non-human primates, we are extending these in vitro models to a diverse set of mammals, thereby enabling comparative embryology in a dish. This broader cross-species perspective allows us to investigate conserved and lineage-specific programs across mammals, further deepening our understanding of the evolutionary principles shaping primate and human development. Through this work, we aim to contribute to the emergence of an exciting new research field: synthetic evolutionary developmental biology.
Ethical Collaboration
Human and non-human primate embryo research has long been riddled with ethical controversy. At the Alev Lab, we work closely with bioethicists to ensure that every aspect of our research is ethically sound. Beyond our own research, we actively engage in the broader conversation on embryo research. We hope to contribute to the development of ethical frameworks, both in Japan and internationally, that support responsible innovation in this rapidly evolving field.
Publications
Matsuda M, Yamanaka Y, Uemura M, Osawa M, Saito MK, Nagahashi A, Nishio M, Guo L, Ikegawa S, Sakurai S, Kihara S, Maurissen TL, Nakamura M, Matsumoto T, Yoshitomi H, Ikeya M, Kawakami N, Yamamoto T, Woltjen K, Ebisuya M, Toguchida J, Alev C. Recapitulating the human segmentation clock with pluripotent stem cells. Nature. 2020 Apr;580(7801):124-129. https://doi.org/10.1038/s41586-020-2144-9
Matsuda M, Hayashi H, Garcia-Ojalvo J, Yoshioka-Kobayashi K, Kageyama R,Yamanaka Y, Ikeya M, Toguchida J, Alev C, Ebisuya M. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science. 2020 Sep 18;369(6510):1450-1455. https://doi.org/10.1126/science.aba7668
Kishimoto K, Furukawa KT, Luz-Madrigal A, Yamaoka A, Matsuoka C, Habu M, Alev C, Zorn AM, Morimoto M. Bidirectional Wnt signaling between endoderm and mesoderm confers tracheal identity in mouse and human cells. Nature Communications. 2020 Aug 27;11(1):4159. https://doi.org/10.1038/s41467-020-17969-w
Hamidi S, Nakaya Y, Nagai H, Alev C, Kasukawa T, Chhabra S, Lee R, Niwa H, Warmflash A, Shibata T, Sheng G. Mesenchymal-epithelial transition regulates initiation of pluripotency exit before gastrulation. Development. 2020 Feb 3;147(3):dev184960. https://doi.org/10.1242/dev.184960
Kawai S, Yoshitomi H, Sunaga J, Alev C, Nagata S, Nishio M, Hada M, Koyama Y, Uemura M, Sekiguchi K, Maekawa H, Ikeya M, Tamaki S, Jin Y, Harada Y, Fukiage K, Adachi T, Matsuda S, Toguchida J. In vitro bone-like nodules generated from patient-derived iPSCs recapitulate pathological bone phenotypes. Nature Biomedical Engineering. 2019 Jul;3(7):558-570. https://doi.org/10.1038/s41551-019-0410-7
McIntyre BA, Alev C, Mechael R, Wu Y, Sheng G, Bhatia M et al. Expansive generation of functional airway epithelium from human embryonic stem cells. Stem Cells Translational Medicine. 2014 Jan; 3(1):7-17. https://doi.org/10.5966/sctm.2013-0119
Alev C, Nakano M, Wu Y, Horiuchi H, Sheng G. Manipulating the avian epiblast and epiblast-derived stem cells. Methods in Molecular Biology. 2013;1074:151-73. https://doi.org/10.1007/978-1-62703-628-3_12
Alev C, Wu Y, Nakaya Y, Sheng G. Decoupling of amniote gastrulation and streak formation reveals a morphogenetic unity in vertebrate mesoderm induction. Development. 2013 Jul;140(13):2691-6. https://doi.org/10.1242/dev.094318
Alev C, Wu Y, Kasukawa T, Jakt LM, Ueda HR, Sheng G. Transcriptomic landscape of the primitive streak. Development. 2010 Sep 1;137(17):2863-74. https://doi.org/10.1242/dev.053462


