Research Overview
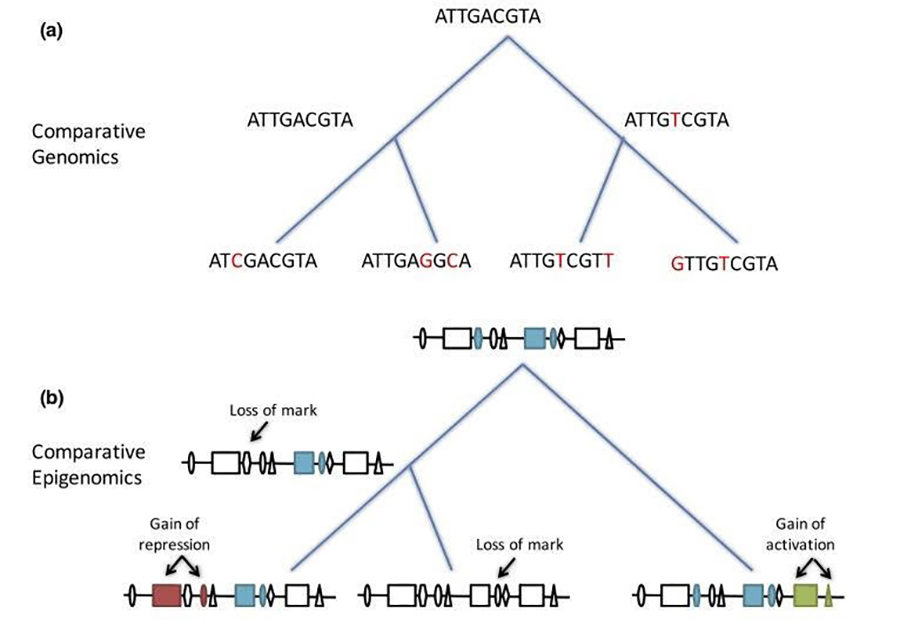
We seek to understand transcription regulation in mammals and characterize the contribution of non-coding DNA variants to human disease. With consortiums such as ENCODE and the International Human Epigenome Consortium (IHEC), the number of datasets characterizing the epigenomic state of different cell types and conditions has been steadily growing. Nonetheless, studies trying to infer the impact of non-coding DNA variants have had to rely on limited functional datasets to train their model and make predictions, which has proven difficult. We have shown that by using an evolutionary perspective to interpret functional genomic datasets, we can better decrypt the role of non-coding human DNA. To improve on this even further, we now propose to build comprehensive epigenomic profiles of specific cell-types in multiple human individuals and in various non-human primate species (including chimpanzee, bonobo, gorilla, rhesus and marmoset). We will then use these datasets profiling intra and inter-species epigenomic variation, to develop and train algorithms to predict the impact of non-coding DNA variants in unprecedented ways (Figure 1).
Areas of interest include: the evolution of regulatory sequences, the role of transposable elements in gene regulation and the impact of genome rearrangements in evolution and cancer. One objective is to develop computational methods and resources for the functional annotation of genomes with a special emphasis on pangenome based approaches for sequencing-based assays (e.g. ChIP-seq, RNA-Seq, exome- and whole-genome sequencing, single-cell analysis).
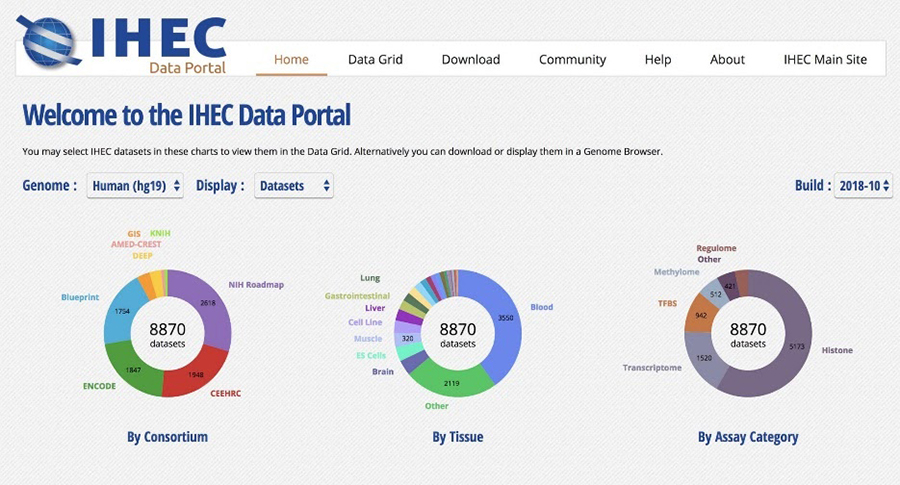
We are also involved in the Global Alliance for Genomics and Health (GA4GH) and develop tools which facilitates data sharing (Figure 2).
Publications
Groza, C., Ge, B., Cheung, W., Pastinen, T., & Bourque, G. (2025). Expanded methylome and quantitative trait loci detection by long-read profiling of personal DNA. Genome Research. https://www.genome.org/cgi/doi/10.1101/gr.279240.124
Aracena, K. A., Lin, Y. L., Luo, K., Pacis, A., Gona, S., Mu, Z., Yotova, V., Sindeaux, R., Pramatarova, A., Simon, M. M., Chen, X., Groza, C., Lougheed, D., Gregoire, R., Brownlee, D., Boye, C., Pique-Regi, R., Li, Y., He, X., Bujold, D., Pastinen, T., Bourque, G., & Barreiro, L. B.(2024). Epigenetic variation impacts individual differences in the transcriptional response to influenza infection. Nature Genetics, 56(3). https://doi.org/10.1038/s41588-024-01668-z
Groza, C., Schwendinger-Schreck, C., Cheung, W. A., Farrow, E. G., Thiffault, I., Lake, J., Rizzo, W. B., Evrony, G., Curran, T., Bourque, G., & Pastinen, T. (2024). Pangenome graphs improve the analysis of structural variants in rare genetic diseases. Nature Communications, 15(1), Article ARTN 657. https://doi.org/10.1038/s41467-024-44980-2
Chen, X., Pacis, A., Aracena, K. A., Gona, S., Kwan, T., Groza, C., Lin, Y. L., Sindeaux, R., Yotova, V., Pramatarova, A., Simon, M. M., Pastinen, T., Barreiro, L. B., & Bourque, G. (2023). Transposable elements are associated with the variable response to influenza infection. Cell Genomics, 3(5), Article ARTN 100292. https://doi.org/10.1016/j.xgen.2023.100292
Groza, C., Chen, X., Pacis, A., Simon, M. M., Pramatarova, A., Aracena, K. A., Pastinen, T., Barreiro, L. B., & Bourque, G. (2023). Genome graphs detect human polymorphisms in active epigenomic state during influenza infection. Cell Genomics, 3(5), Article ARTN 100294. https://doi.org/10.1016/j.xgen.2023.100294
Groza, C., Kwan, T., Soranzo, N., Pastinen, T., & Bourque, G. (2020). Personalized and graph genomes reveal missing signal in epigenomic data. Genome Biology, 21(1), Article ARTN 124.
https://doi.org/10.1186/s13059-020-02038-8
Bogdan, L., Barreiro, L., & Bourque, G. (2020). Transposable elements have contributed human regulatory regions that are activated upon bacterial infection. Philosophical Transactions of the Royal Society B: Biological Sciences, 375(1795), Article ARTN 20190332. https://doi.org/10.1098/rstb.2019.0332
Bourque, G., Burns, K. H., Gehring, M., Gorbunova, V., Seluanov, A., Hammell, M., Imbeault, M., Izsvák, Z., Levin, H. L., Macfarlan, T. S., Mager, D. L., & Feschotte, C. (2018). Ten things you should know about transposable elements. Genome Biology, 19, Article ARTN 199. https://doi.org/10.1186/s13059-018-1577-z
Bujold, D., Morais, D., Gauthier, C., Côté, C., Caron, M., Kwan, T.,…Bourque, G. (2016). The International Human Epigenome Consortium Data Portal. Cell Systems, 3(5), 496-+. https://doi.org/10.1016/j.cels.2016.10.019