RECODE

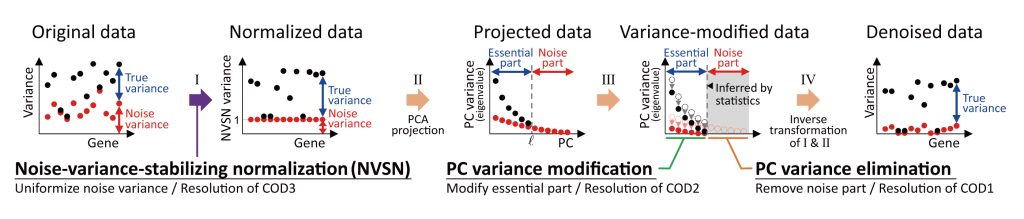
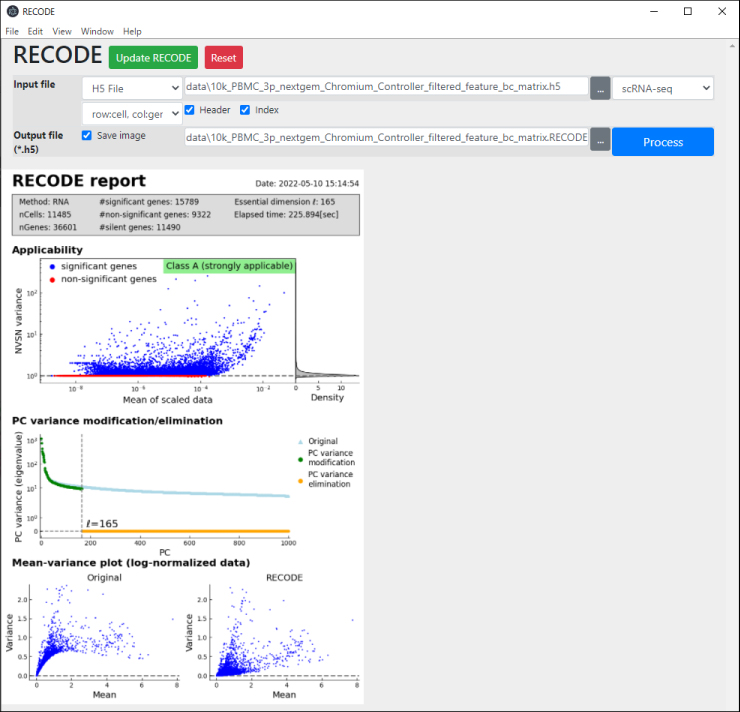
Resolution of the curse of dimensionality (RECODE) is a noise reduction method for single-cell sequencing data based on high-dimensional statistics.
The license gives permission for personal, academic, or educational use. Any commercial use is strictly prohibited. Please contact imoto.yusuke.4e<at>kyoto-u.ac.jp for licensing terms for any commercial use.
Python code
Installation
To install RECODE package, use pip as follows:
$ pip install screcodePyPi downloads (by PePy)



Documentation
Requirements
- Python3
- numpy
- scipy
- scikit-learn
R code
Remark: The current version of the R code is not fast because of the lower speed of the PCA algorithm on R. Therefore, we recommend using the python code for large-scale data.
Installation
You can install RECODE on R with:
devtools::install_github("yusuke-imoto-lab/RECODE/R")Tutorials
For the single-cell sequeincing data X (rows: genes/epigenomes, columns: cells), we can apply RECODE as follows.
library(RECODE)
X.RECODE <- RECODE(X)In the Seurat analysis, we can apply RECODE to SeuratObject and set it as default, as follows:
library(RECODE)
library(Matrix)
data <- as.matrix(seurat[["RNA"]]@counts)
data_RECODE <- RECODE(data)
seurat[["RECODE"]] <- CreateAssayObject(counts = Matrix(data_RECODE, sparse = TRUE))
DefaultAssay(seurat) <- "RECODE"For a detailed analysis, please see below:
Tutorial (Run, QC, Clustering, Annotating etc.)
R code (Python calling)
Installation
After installing remotes (install.packages("remotes")), you can install “recodeinstaller” with the following command:
remotes::install_github("yusuke-imoto-lab/recodeinstaller")Then, the following command installs the Python version of RECODE.
library(recodeinstaller)
install_screcode()Regarding the detail of installer, please see recodeinstaller.
Tutorials
For the single-cell sequeincing data X (rows: genes/epigenomes, columns: cells), we can apply RECODE as follows.
library(reticulate)
source("recodeloader/load_recodeenv.R")
plt <- reticulate::import(module="matplotlib.pyplot")
screcode <- reticulate::import(module="screcode.screcode")
X.RECODE<-screcode$RECODE(X)Below is a more detailed analysis:
Desktop Application
Windows (exe) and MAC OS (dmg) applications are available.